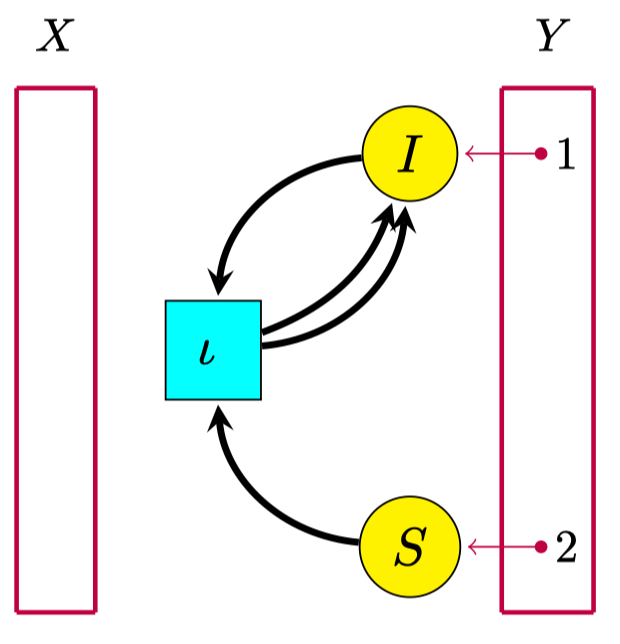
Reaction network (S,T,s,t)
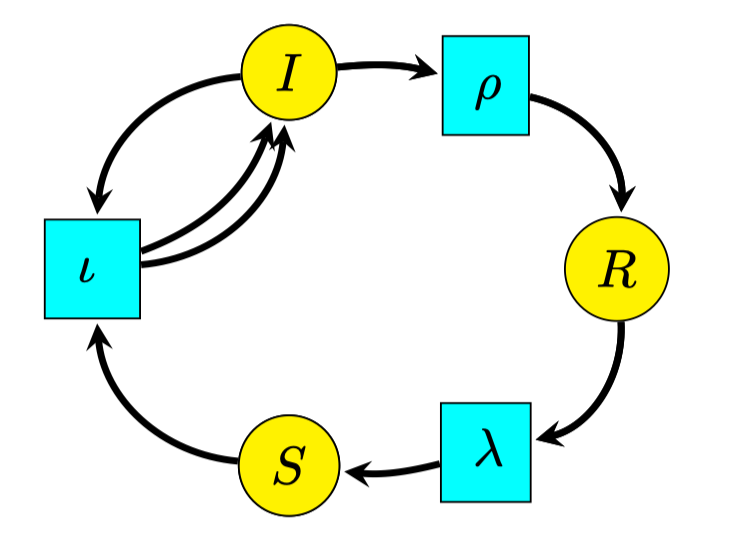
- Eg: SIRS infectious disease model
- S+Iι2I
- IρRλS
- species:
- S: susceptible
- I: infected
- R: resistant
- reactions:
- ι: infection
- ρ: recovery
- λ: loss of resistance
- definition:
- a finite set of species
- reactions go between “complexes”, which is finite linear combinations of these species with natural number coefficients
- rate constant and rate equation:
- rate constant: a positive number called can be attached to each reaction
- rate equation: now a reaction network determines a system of differential equations saying how the concentrations of the species change over time
- Eg
- dtdS=rλR−rιSI
- dtdI=rιSI−rρI
- dtdR =rρI−rλR
- interesting properties:
- existence and uniqueness of steady state solutions
- Mathematics definition:
- consists of:
- a finite set S
- : elements are called species
- a finite set T
- : elements are called transitions
- functions s,t:T→NS
- NS’s elements are called complexes, which is finite linear combinations of these species with natural number coefficients.
- any transition τ∈T has a source s(τ) and a target t(τ)
- if s(τ)=κ and t(τ)=κ′, we write τ:κ→κ′
- the set of complexes relevant to a given reaction network is
- : K=im(s)∪im(t)⊆NS
- graph:
- a reaction network gives a graph
- : vertices set of K
- : an edge for each transition τ:κ→κ′
- it can have multiple edges or self-loops, thus sometimes called:
- a directed multigraph
- or a quiver
Petri Net
- bipartite directed graph
- : 2 kinds of vertices, species and reactions
- : edges
- into a reaction, specifying its input
- out, specifying its output
- Terminology
- in Petri net literature, species are called “places”, and reactions are called “transitions”
- so Petri net is sometimes called place-transition net or P/T net
- stochastic Petri net: when each reaction has a rate constant attached
- open Petri net
- inputs and outputs: species can flow in or out
- open rate equation: the usual one with extra terms describing inflows and outflows